Overview
Our platform enables the rapid annotation of medical terms within clinical notes. A user can highlight spans of text and quickly map them to concepts in large vocabularies within a single, intuitive platform. Users can use the search and recommendation features to find labels without ever needing to leave the interface. Further, the platform can take in output from existing clinical concept extraction systems as pre-annotations, which users can accept or modify in a single click. These features allow users to focus their time and energy on harder examples instead. See the demo below!
Why?
Clinical notes document detailed patient information, including pertinent medical history, procedures performed during a visit, and response to treatment. This information can be leveraged for a variety of use cases–from more personalized decision support in electronic health record to richer retrospective research.
However, this data often exists only in clinical text, and not in any structured fields; therefore, many use cases involve manual or automated structuring of relevant concepts from clinical notes. Current automated approaches of extracting structured clinical concepts are insufficiently robust for many downstream uses, and there isn’t much annotated training data publicly available to improve machine learning models. The goal of our platform is to make that annotation process easier and faster: whether for a manual curation use case, or for the creation of an annotated training dataset.
Features
Pre-Annotations
Sometimes concept mentions are simple and straightforward for algorithms to recognize. In these cases, pre-annotations can save annotators time and energy. Pre-annotations outline the suggested span of text alongside its predicted label. The user can then to choose to accept the machine suggestion in just a single click, or modify or delete. In the image below, there are 3 pre-annotations: on flagyl, BUN, and NGT.

PRAnCER can flexibly set the pre-annotations to the outputs of any clinical entity normalization system (MetaMap, cTAKES, ClinicalBERT). A user just provides the spans and expected labels in a CSV file. Alternately, a user can provide a dictionary from text to the CUI they want it to pre-fill to; a sample dictionary is provided.
Recommendations
Even when a model can’t settle on a single label with high-confidence, it can often surface a correct label in its top few predictions. PRAnCER comes built-in with an NLP recommendation algorithm for suggesting likely concept labels once a span of text is highlighted. Below you can see that we can correctly recommend ‘vancomycin’ for the highlighted vanco. The recommendation function is merely a Python call, so one can easily swap out our recommendation algorithm for any new model.

Search
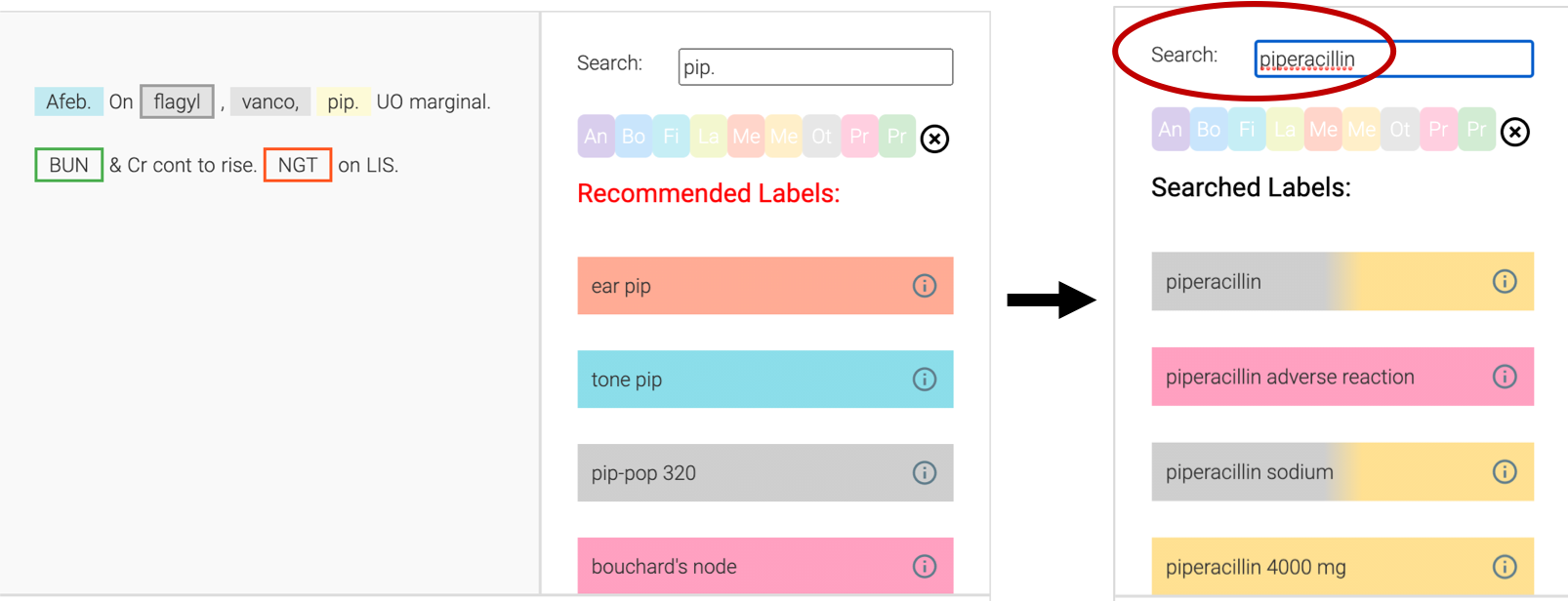
When the recommendations fail to bring up the correct label(s), the annotator can easily search for their desired term without having to leave the interface. In the example below, the annotator could search ‘piperacillin’ and directly click to select.
UMLS Linking
Medical ontologies are large and nuanced, and sometimes it is necessary to get more information before choosing a concept label. You can link your UMLS account to our interface, and as shown below, it will automatically surface the UMLS-provided definitions for a concept in one click.

Concept Categories
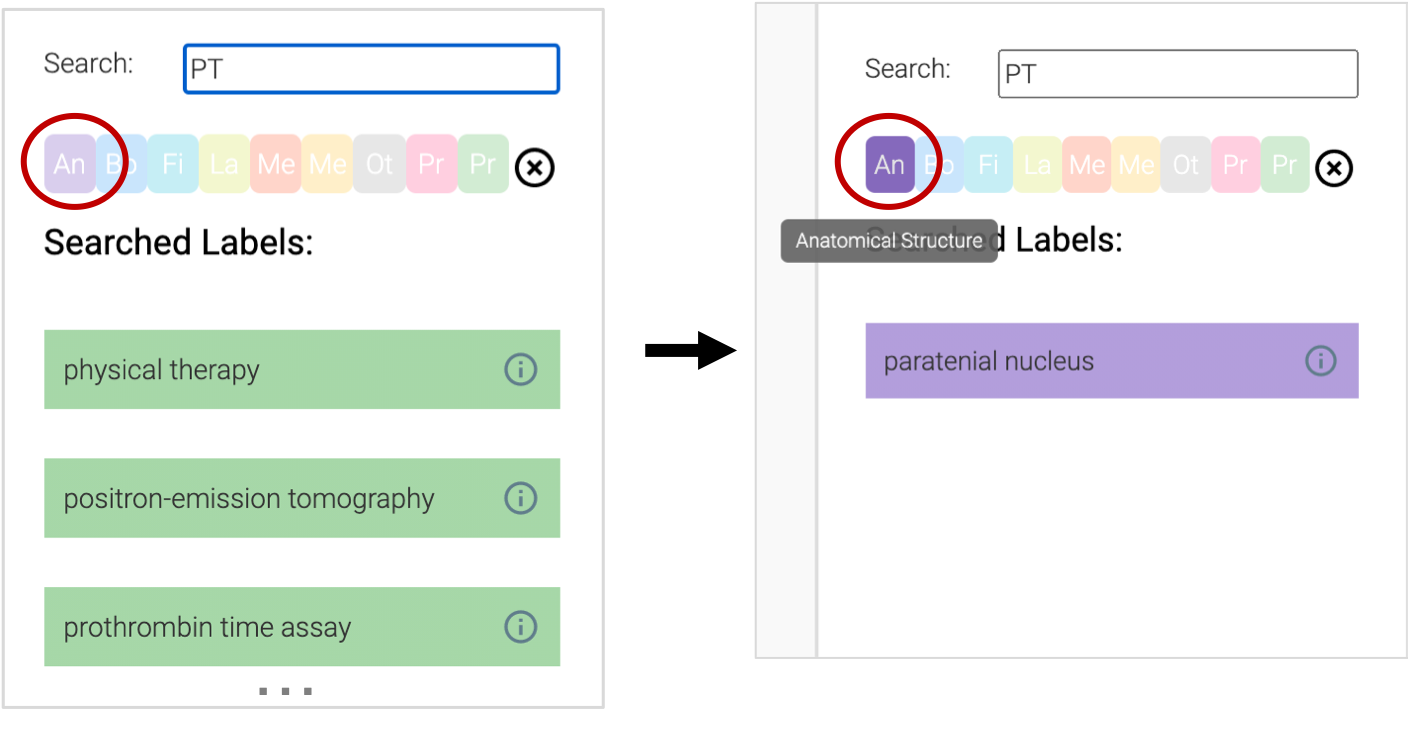
We provide a color-coding for each concept type, e.g. findings, procedures, medical devices; these can be used as visual cues to quickly find the desired concept label. Further, users can choose to filter concepts by these concept types; below, the user chose to filter the ‘PT’ terms to anatomical terms.
Flexible Annotation
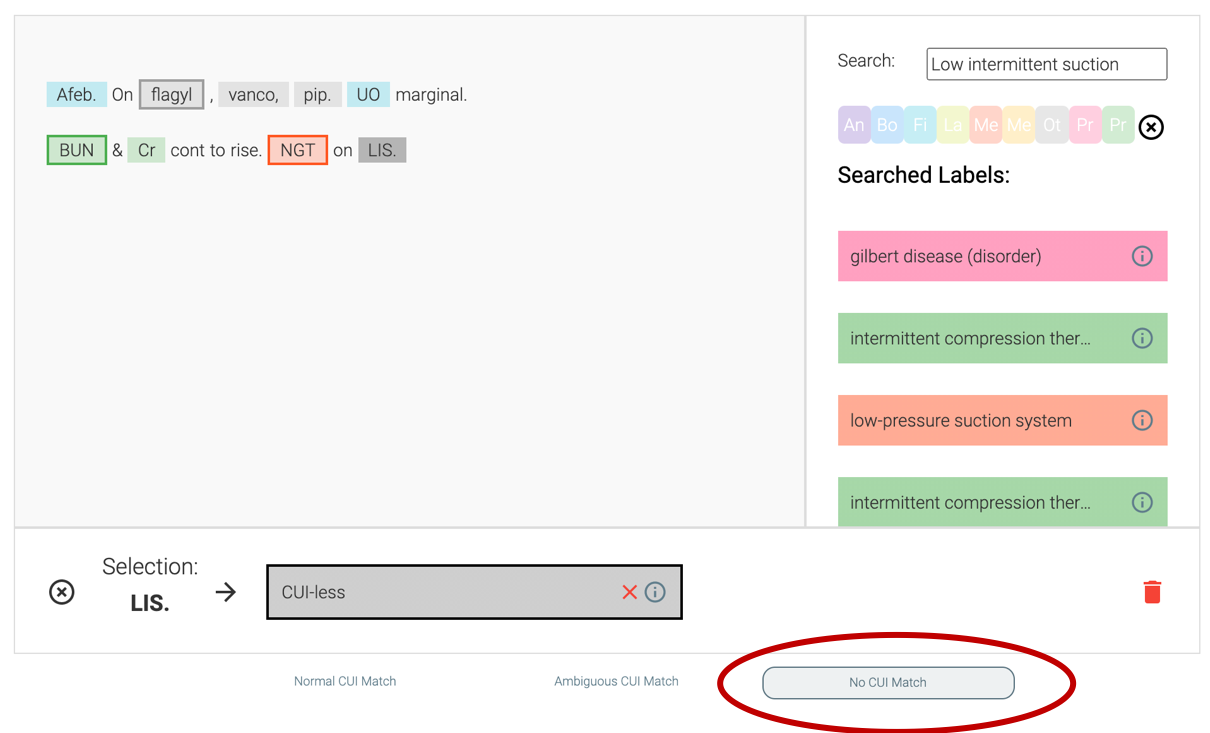
We understand that annotation is often messy and inexact, so PRAnCER provide tooling that can help support such use cases. Users can choose multiple concepts, label a concept as an ‘ambiguous match’, or as below, indicate there was no direct match found. PRAnCER also enables the annotation of overlapping spans.
How to Use
Installation Instructions
PRAncer is built on python3 and Node.Js; the README of our Github provides detailed instructions on how to install PRAnCER on your machine in a few simple steps. PRAnCER can operate on Mac, Windows, and Linux machines.
Loading in Data
To load in data, users directly place any clinical notes as .txt files in the /data folder of our annotaor. If the user wants to load in pre-annotations, users can place a CSV of spans and CUIs, and our scripts will automatically incorporate those annotations. An example .txt file and .csv file are provided.
UMLS Vocabulary
Use of the platform requires a UMLS license, as it requires several UMLS-derived files to surface recommendations, as well as a UMLS API key to surface additional information. Please email magrawal (at) mit (dot) edu to request these files, along with your API key so we may confirm. You can sign up here.